By Katherine McFerrin

We are excited to announce that we have characterized a novel henipavirus, Angavokely Virus (AngV). Our recent publication in the Journal of Virology describes the complete coding sequence of AngV from urine samples of the Malagasy fruit bat, Eidolon dupreanum. In prior work AngV had only been identified by serology.
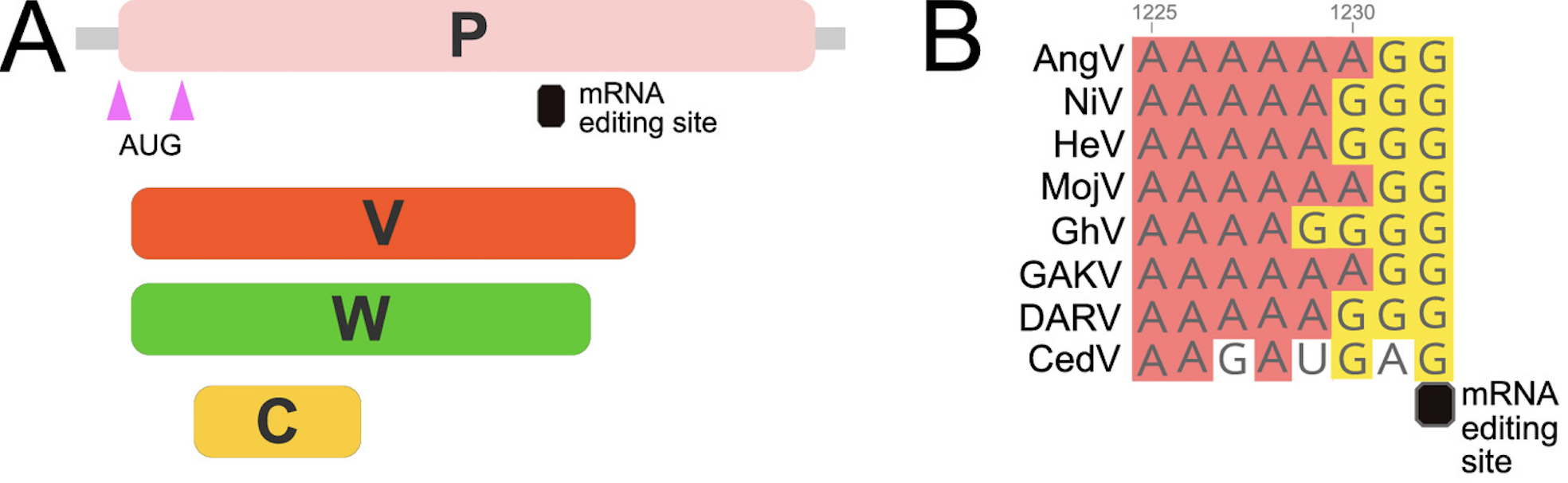
Henipaviruses are highly pathogenic emerging zoonotic viruses from bat, rodent and shrew reservoirs. AngV may fall within the ranks of other pathogenic henipaviruses including Hendra and Nipah virus following spillover to human hosts. Our work reveals that AngV encodes major proteins associated with pathogenic henipaviruses, notably, the phosphoprotein (P-protein) embedded V and W proteins, which are known to evoke host immune responses. V and W proteins in pathogenic Hendra (HeV), Nipah (NiV), Mojiang (MojV), Daeryong (DARV) and Gamak viruses (GAKV) and Ghanian bat Henipavirus (GhV), but are absent in non-pathogenic Cedar virus (CedV). This suggests that AngV is likely also pathogenic.
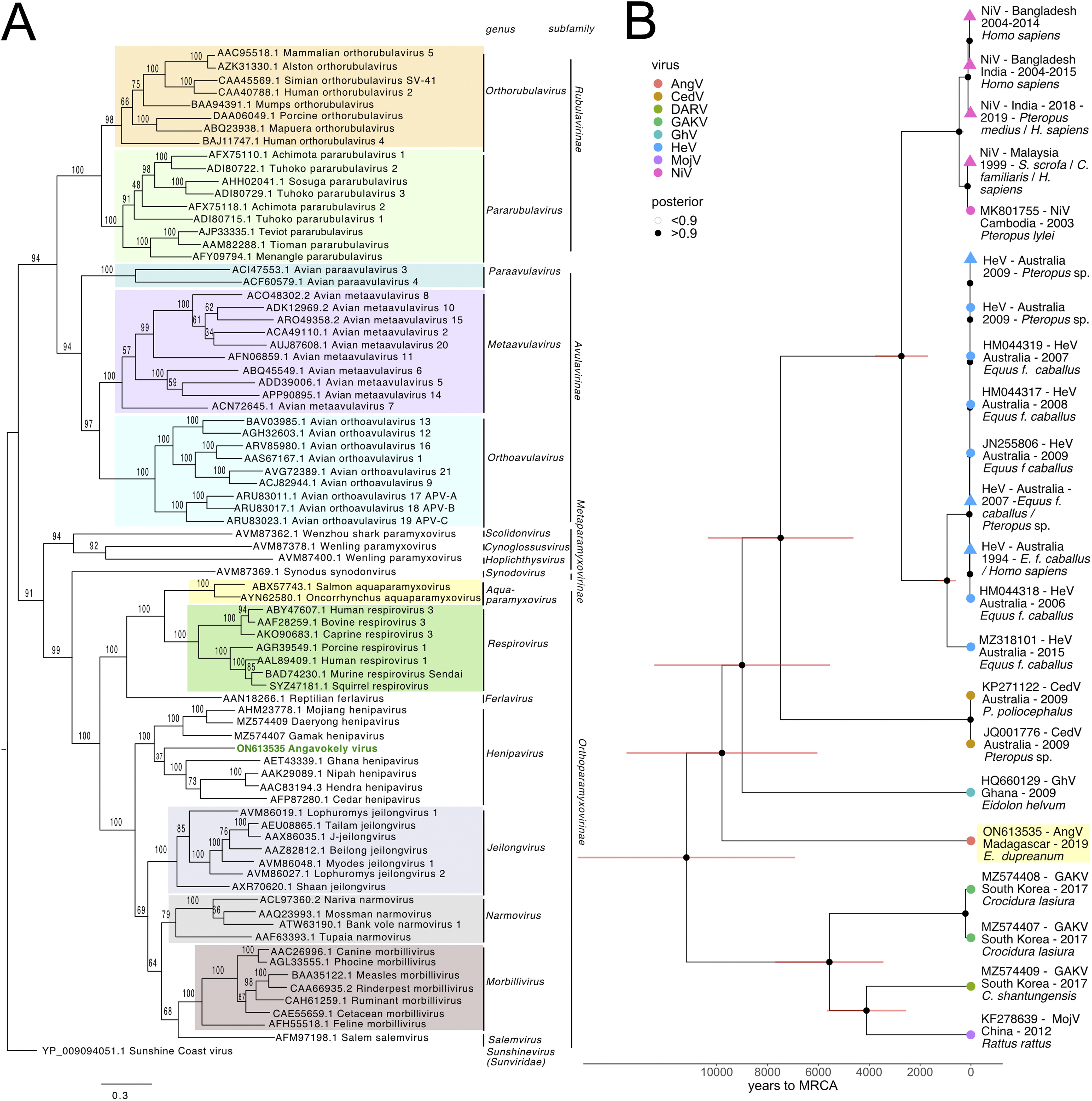
AngV is an ancestral bat-borne henipavirus. Using maximum-likelihood and Bayesian phylogenetic analysis we show that AngV is ancestral to the 4 previously described bat-borne henipaviruses (NiV, HeV, CedV, and GhV) but more recently evolved than the rodent and shrew-borne clade of MojV, GAKV, and DARV.
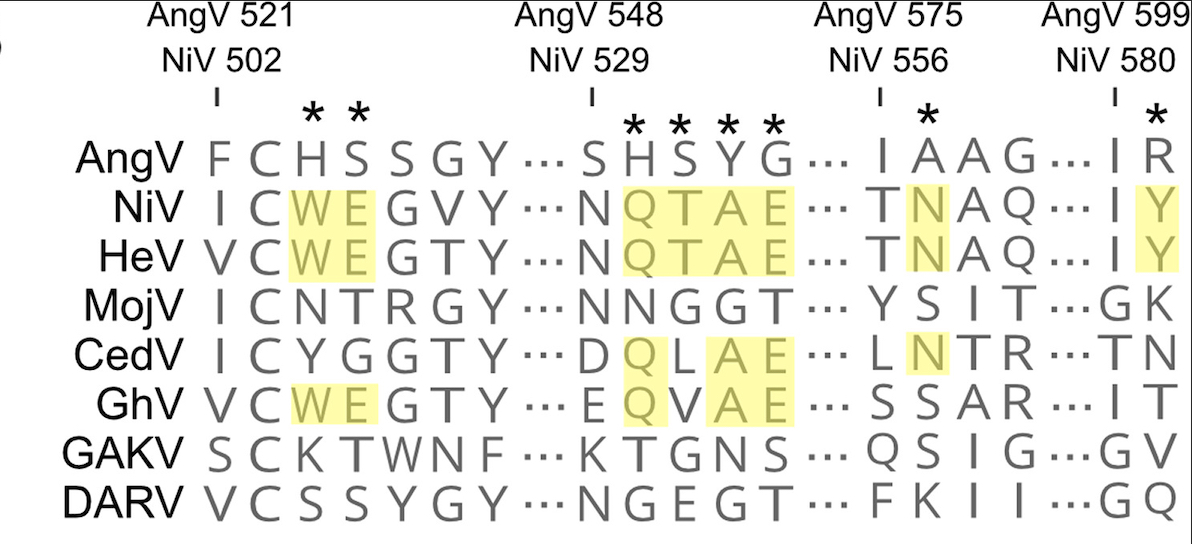
AngV also provides insights into the greater evolutionary story of henipavirus pathogenesis. AngV adds to the growing number of novel henipaviruses that do not use an ephrin binding cellular entry mechanism. Ephrin type A and B receptor binding allows most Henipaviruses to gain cell entry. Receptors for the MojV clade and AngV, however, remain unknown. AngV lacks the well conserved ephrin-binding residues, F and G proteins, found in other henipaviruses, leaving the door open for novel cell entry mechanisms.
Overall, the discovery and characterization of AngV emphasizes the importance of viral survelliance in Madagascar’s bat populations for circulating henipaviruses. Bats are hunted and consumed as a food source in Madagascar, increasing the chance of disease spillover. Additionally, 9 out of 45 Malagasy bat species can be found across Africa, Asia and/or Europe, which highlights the potential for island hopping to facilitate inter-species viral transmission. Future work to decipher AngV’s cell entry mechanism will allow us to determine the range of hosts for the virus and its zoonotic potential.